We are increasingly moving towards data-sharing and the development of online databases in ecology. Any scientist today can access trait data for thousands of species, global range maps, gene sequences, population time series, or fossil measurements. Regardless of arguments for or against, the fact that massive amounts of ecological data are widely available is changing how research is done.
For example, global trait databases (TRY is probably best known) allow researchers to explore trait-based measures in communities, habitats, or ecosystems without requiring that the researchers have actually measured the traits of interest in the field. And while few researchers would suggest that this is superior to making the measurements in situ, the reality is that there are many situations where trait data might be required without the researcher being able to make them. In these cases, online databases are like a one-stop shop for data. But despite the increasing frequency of citations for trait databases, until now there has been little attempt to quantify how well database values act as proxies for observed trait values. How much should we be relying on these databases?
There are many well-recorded reasons why an average trait value might differ from an individual value: intraspecific differences result from plasticity, genotype differences, and age or stage differences, all of which may vary meaningfully between habitats. How much this variation actually matters to trait-based questions is still up for debate, but clearly affects the value of such databases. To look at this question, Cordlandwehr et al. (2013) examined how average trait values calculated with values from a North-west European trait database (LEDA) corresponded with average trait values calculated using in situ measurements. Average trait values were calculated across several spatial scales and habitat types. The authors looked plant communities growing in 70 2m x 2m plots in the Netherlands, divided between wet meadow and salt marsh habitats. In each community, they measure three very common plant traits: canopy height (CH), leaf dry matter content (LDMC), and specific leaf area (SLA).
In situ measurements were made such that the trait value for a given plot was the median value of all individuals measured; for each habitat it was the the median value of all individuals measured in the habitat. The authors calculated the average trait values (weighted by species abundance) across all species for each community (2m x 2m plot) and each habitat (wet meadow vs. salt marsh). They then compared the community or habitat average as calculated using the in situ values and the regional database values.
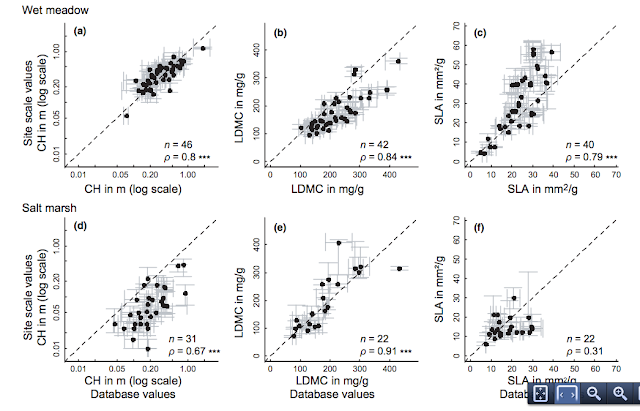 | ||
| From Cordlandwehr et al. 2013. Habitat-level traits at site scale plotted against habitat-level traits calculated using trait values retrieved from a database. |
The authors found the correspondence between average trait values measured using in situ or database values varied with the scale of aggregation, the type of trait and the particular habitat. For example, leaf dry matter content varied very little but SLA was variable. The mesic habitat (wet meadow) was easier to predict from database values than the salt marsh habitat, probably because salt marshes are stressful environments likely to impose a strong environmental filter on individuals, so that trait values are biased. While true that rank differences in species trait values tended to be maintained regardless of the source of data, intraspecific variation was high enough to lead to over- or under-prediction when database values were relied on. Most importantly, spatial scale mattered a lot. In general, database values at the habitat-scale were reasonable predictors of observed traits. However, the authors strongly cautioned against scaling such database values to the community level or indeed using averaged values of any type at that scale: “From the poor correspondence of community-level traits with respect to within-community trait variability, we conclude that neither average trait values of species measured at the site scale nor those retrieved from a database can be used to study processes operating at the plot scale, such as niche partitioning and competitive exclusion. For these questions, it is strongly recommended to rigorously sample individual plants at the plot scale to calculate functional traits per species and community.”
There are two conclusions I take from this. First, that the correlation between sampling effort and payoff is still (as usual) high. It may be easier to get traits from a database, but it is not usually better. The second is that studies like this allow us to find a middle ground between unquestioning acceptance or automatic criticism of trait databases: they help scientists develop a nuanced view that acknowledges both strengths and weaknesses. And that's a valuable contribution for a study to make.

No comments:
Post a Comment